Methodology
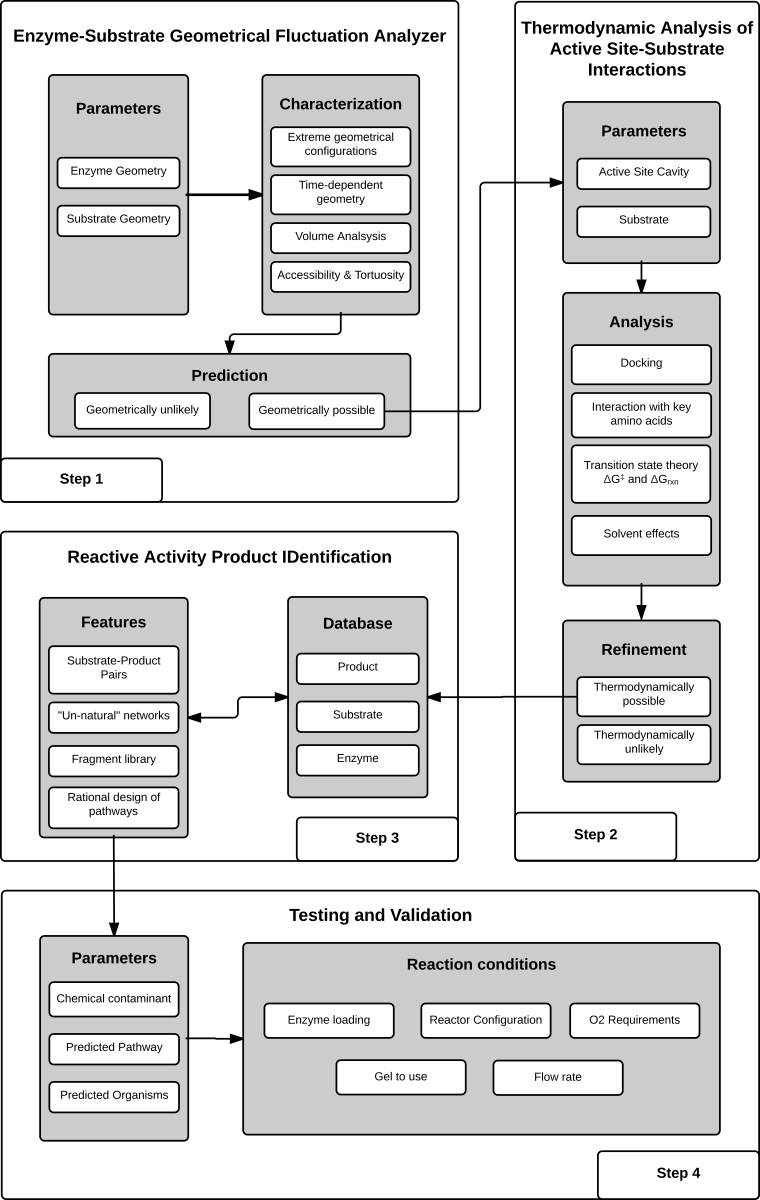
The RAPID algorithm is an integrated set of analyses used to generate a prediction on the probability that a compound is a substrate, or non-substrate of an enzyme.
Step 1: In order for a chemical to be a substrate it has to fulfil the condition of reaching the enzyme's active site, as that is the reactive center. The process of entering the active is modeled by RAPID as a molecule flowing through a tortuos channel. The irregular architecture of the channel provides a means for hindering certain molecules from reaching the active site. This hindrances can be caused by simple geometric constraints, or can be more complex thus requiring favorable electrostatic interactions between the small molecule and the enzyme.
Step 2: If the small molecule meets the condition of reaching the active site, then a favorable positioning needs to be attained. If the molecule is able to adopt a conformation that is to prone to chemical attack by the enzyme then the probabilty of being a substarte increases. In addition, for certain molecules it is a necessary condition that interactions with specific amino acids are present. These types of interactions can include, but are not limited to, hydrogen bonding and, π-π stacking.
Step 3: If the entrance (step 1) and active site (step 2) conditions both met, then the substrate is classified as highly likely to be a substrate for a given enzyme and thus stored into the RAPID prediction database. In addition, there are certain enzymes which carry out a known reaction, for instance dioxygenases (see Naphthalene 1,2-dioxygenase), and thus a possible product of the reaction can also be predicted.
Step 4: Once a prediction has been made, it is up to the user to validate the prediction through experiments. We ask that if you provide feedback at rapid@umn.edu if you have tested one of our predictions. In this way we can continually improve the quality of the predictions made.